Areas of investigation/research focus
The main focus of my group is on dissecting the relationship between interchromatin space, chromatin and transcription, and how the underlying molecular mechanisms coordinate stress response in age-related processes and neurodegeneration. Our work carries multiple implications ranging from understanding disease etiology to the identification of druggable targets. Our studies comprise mouse models, patient-derived material and state-of-the art (epi)genomics, the latter supported by strong bioinformatics/computational biology core.
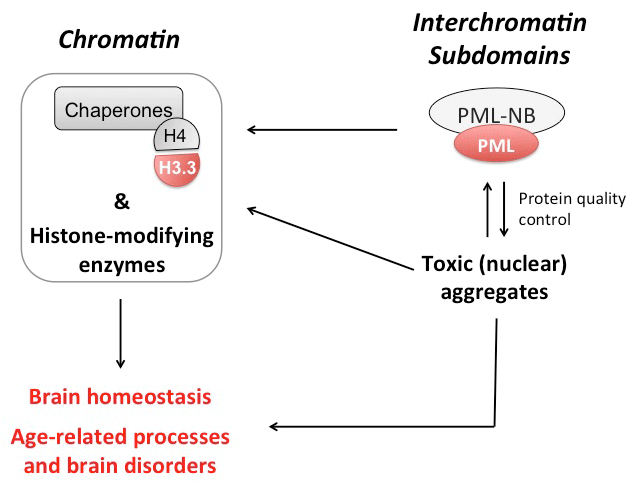
The PML nuclear body in brain homeostasis and stress response. The Promyelocytic Leukaemia (PML) scaffold protein is the essential component of a subnuclear structure of the interchromatin space called the PML-nuclear body (PML-NB) and implicated in cell fate control in physiological settings and upon stress. PML regulates stem cell fate during brain development (Regad et al, Nat Neurosci 2009), with potential implications for cognitive function and behaviour. Furthermore, PML controls cell migration during neurogenesis via epigenetic control of axon guidance genes (Amodeo, A et al, Cell Rep 2017). PML represses transcription via regulation of the Polycomb Repressive Complex 2 protein levels. As PML mediates proteostasis of misfolded proteins and protection against neurodegeneration, we are studying PML role in nuclear protein quality control and stability in neurons with a particular focus on chromatin-remodelling complexes (see below) in normal physiological settings and in neurodegenerative disease models.
Epigenetic control in the onset and progression of age-related disorders/pathologies
The PML-interacting DAXX and ATRX proteins mediate deposition of the histone H3.3 variant, which plays a critical role in postmitotic cells due to its replication-independent loading. We showed that calcium signalling upon neuronal activation promotes H3.3 loading for chromatin remodelling and transcription in the CNS (Michod et al, Neuron 2012), with implications for neuronal plasticity, depressive-like behaviour and aging. In this respect, Daniele Bano group (DZNE Bonn) in collaboration with us implicated H3.3 loading in lifespan extension mechanisms at organismal level (Piazzesi et al, Cell Rep 2017). Finally, driver mutations in H3.3 as well as in ATRX and DAXX are found in brain cancer. Therefore, H3.3 is a multifaceted epigenetic player at the crossroad of brain plasticity, age-related disorders and neoplastic brain disease.
We are now investigating the role of H3.3 loading and associated histone-modifying enzymes (as well as PML-NBs, see above) in coordinating stereotypic stress response pathways, aging and onset/progression of neurodegenerative conditions. As part of our translational efforts, we are running high-content screens to identify small molecules modulating histone variant loading in both physiological and pathological settings.
Finally, ERC-funded work in our lab has led to generation of a faithful somatic model of mutant H3.3-driven tumorigenesis, providing insights into disease mechanisms (Pathania et al, Cancer Cell 2017; F1000). Our experience in modelling and studying chromatin-associated pathways underlying neoplastic disease places our group in the unique position to compare epigenetic reprogramming in age-related brain disorders and brain cancer, offering us the opportunity to identify potential common mechanisms.