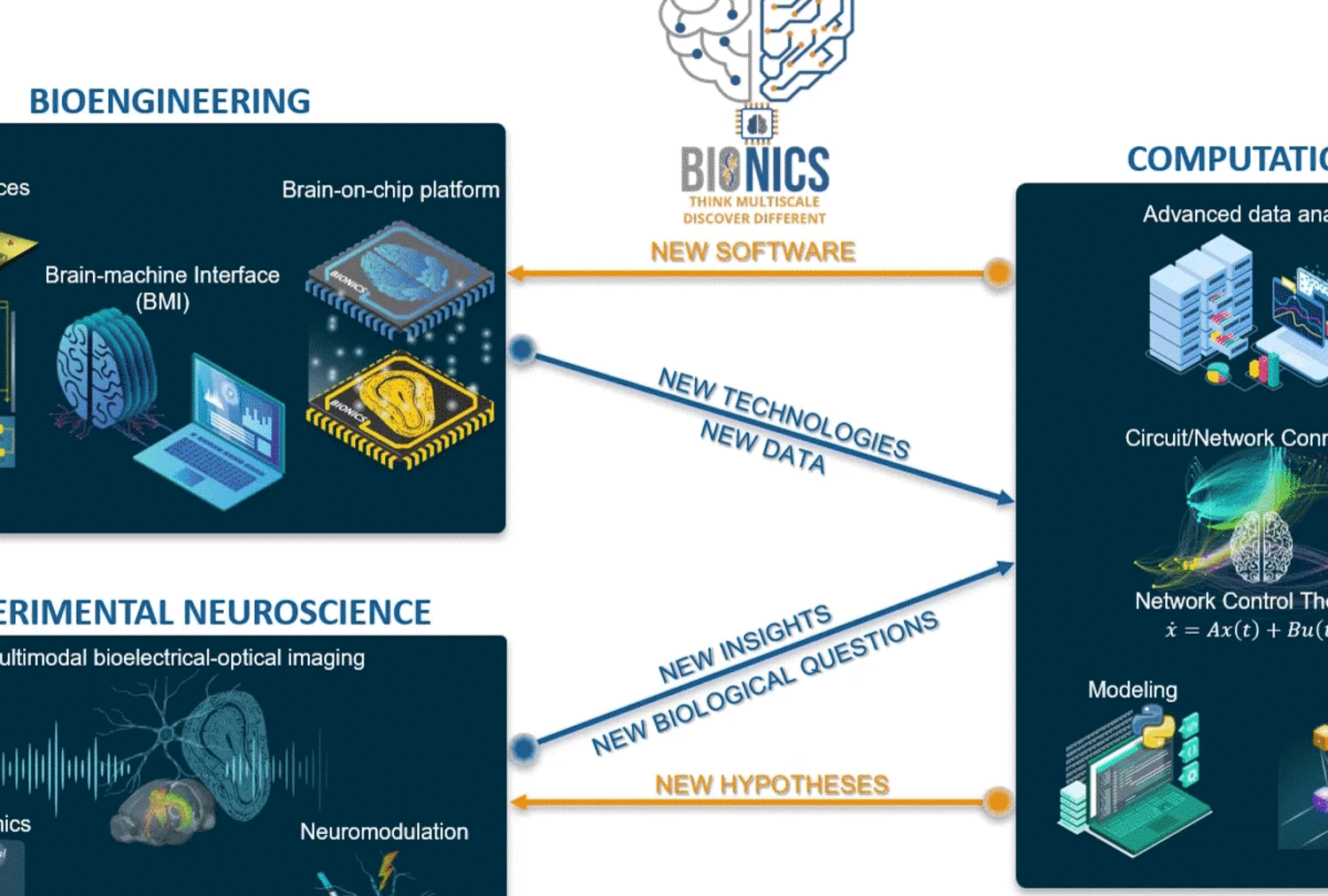
Areas of investigation/research focus
Welcome to BIONICS, where we blend neuroscience and technology to push the limits of what's possible in biomedical engineering. Our interdisciplinary research group is dedicated to exploring the frontiers of science and developing cutting-edge technologies to understand the complex network dynamics of the brain in health and disease states. Using a unique combination of engineering, experimental neuroscience, and computational tools, we aim to decipher how neural computations are coordinated and integrated across scales to generate robust functional outcomes. Our research focuses on identifying the yin and yang of various neural computations performed by different brain regions and the collective network stability during network dynamical changes induced by experience, learning, disease, and neurogenesis.
One of our approaches involves exploiting the reparative regenerative capability of the brain to rewire existing distributed circuitry promoted by adult neurogenesis in the hippocampus. By promoting adult neurogenesis through several experience paradigms, we aim to delay the onset of multiple neurodegenerative diseases and various forms of brain injury. Knowledge of large-scale neural dynamics has a promising future impact. We can create more effective neuroscience-oriented computational models and develop advanced artificial intelligence (AI) frameworks. Furthermore, we can expand the reach of memory neuromorphic brain-inspired sensors and chips into new applications in health and disease.
Our research mission has significant societal impacts by addressing mechanisms for neurodegenerative diseases such as Alzheimer’s disease and advancing novel bioelectronics therapies that could have direct translational targets.
Guided by our unique engineering platforms and computational frameworks, we gather multiscale and multimodal experimental information with possible neural network modulation to provide unique large-scale readouts from genes to functional network levels at high spatiotemporal resolution. Our experimental neuroscience is empowered by novel brain-on-chip technology and methods to enhance neuronal interface for novel readouts. At the same time, we have implemented several computational tools to analyze our multidimensional data and provide a deeper characterization of neuronal communication in circuits and networks using graph analysis, biophysical modeling, control theory, and machine learning tools.
At BIONICS, we are driven by our passion for understanding the complexity of neural computations across scales and linking neuroscience, neurotechnology, and computer science on all levels. We strive to uncover the network deficit mechanisms underlying brain dysfunctions, identify novel targets for developing more efficacious therapeutics, and promote the development of next-generation brain-machine interfaces for restoring and enhancing memory functions impaired in diseases and injuries.
Ongoing Projects
- High-density Biosensor to study coding information in multi-layered Olfactory circuitry
- Assessing hippocampal circuit dynamics in experience-dependent plasticity
- Neurogenic bottom-up computational modeling to elucidate the controllability and criticality of Hippocampal dynamics
- Linking spatial transcriptomics and functional dynamics in large-scale brain network
- Understanding neuronal network complexity for reversible network disintegration - see Project here https://www.dzne.de/en/research/projects/i3d-markers/
- Large-scale biosensor for mapping network-based synaptic transmission and LTP in the Hippocampal circuit.
- Simultaneous optical and electrical recordings of large-scale neural network
Lab Platforms and Techniques
We have implemented interdisciplinary tools to index various neuroscience challenges across scales using the following:
- High-resolution high-density Biosensors
- Calcium imaging
- Patch-clamp recordings
- In-vitro HiPSC-derived neuronal cultures and ex-vivo transgenic mouse brain/olfactory slices
- Genome-wide spatial transcriptomic profiling
- Opto-and-chemogenetic
- Molecular and cellular techniques
- Micro-/nanotechnology
- Computational analytical tools and modeling
Student Opportunities
Attention neuroscience, bioengineering, mathematics, and computer science students! Are you ready to take your studies to the next level? BIONICS lab is seeking highly motivated Bachelor's and Master's students to join our team and work on cutting-edge projects in the field of neurotechnology. With the guidance of experienced professionals, you will gain hands-on experience using the latest computational tools and cutting-edge neurotechnologies. Plus, you will be part of a dynamic and international team of researchers pushing the boundaries of what's possible in the field of neural engineering. Don't miss this opportunity to advance your career and make an impact.
Apply by sending your CV and a cover letter by email to Dr. Hayder Amin (hayder.amin(at)dzne.de)